Expression Pattern Database
The Hope Laboratory Expression Pattern Database is no longer available here. The data from this database in available in WormBase.
A brief introduction to this database
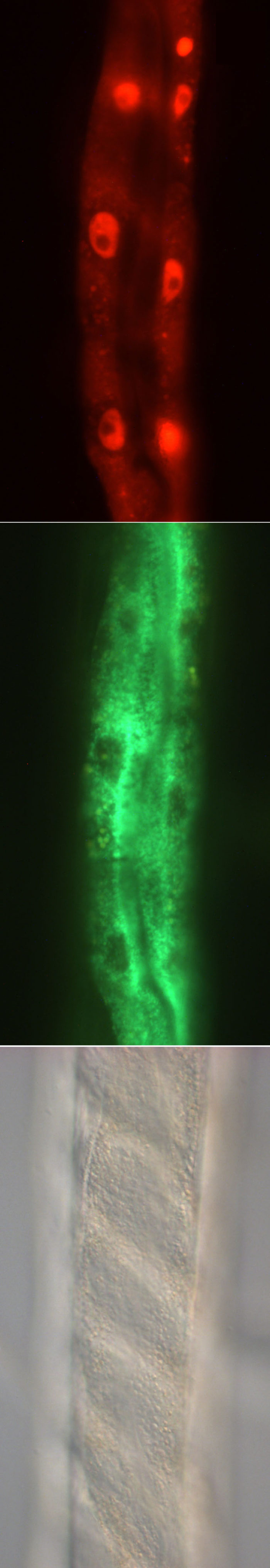
R11A8.4::mCherry and R11A8.5::GFP in strain UL3465 showing expression in the intestine. The two genes are in a single operon.
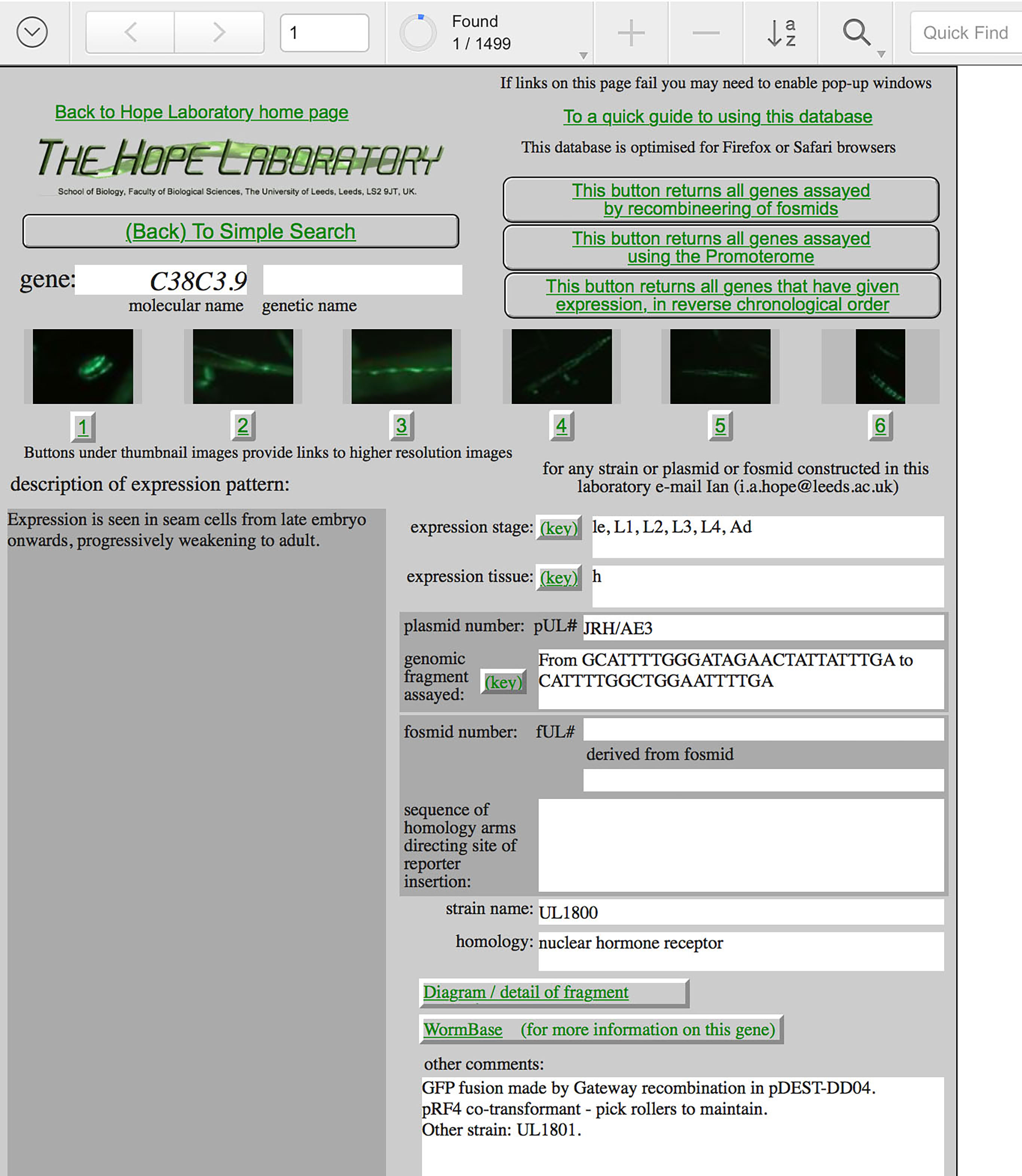
Each page in this database is a record of the results of one experiment. See the example page below. For each experiment, a C. elegans genomic DNA fragment has been joined to a reporter gene, either lacZ or gfp and this fusion gene has been used to generate transgenic C. elegans lines. The expression of the reporter gene can be readily observed in the transgenic animals and reveals when, during the life of the animal, and where, within the animal, the DNA fragment being assayed drives gene expression. Typically, the fragment assayed contains the promoter region of a C. elegans gene, as identified at the top of each record, and the expression pattern observed may correspond to that gene. The observed expression pattern need not accurately reflect that of the gene identified, however, as although most major regulatory elements are thought to reside in promoter regions some elements important for the regulation of the expression of that gene may not be contained in the fragment assayed. Indeed, although each DNA fragment assayed is (or was) predicted to contain the start of a gene, not all drive reporter expression.
A brief text description and some representative micrographs of the expression pattern are provided in each record. For lacZ fusions, staining yields a blue precipitate where the gene is expressed. For gfp fusions, the green fluorescence of the reporter gene product can be observed directly in the live animal with an appropriate microscope. Larger versions of the micrographs can be accessed using the buttons beneath each image in a record. Summaries of the stage of expression within the life-cycle and of the tissues in which expression is observed are provided. Each independently generated transgenic strain, with which the expression pattern was characterized and that has been retained for future use, has a unique name, ULxxxx. The cloned plasmid DNA or fosmid DNAs, which contain each reporter gene fusion assayed, also have a unique name, pUL#xxxx or fUL#xxxx. Any strain, plasmid or fosmid is freely available upon request: e-mail Ian Hope. Precise details of the DNA fragment that has been assayed are provided with reference to the C. elegans genomic DNA sequence as annotated in ACeDB/WormBase. Further information such as on the construction of the reporter gene fusion and on maintenance of the transgenic strain are provided under "other comments".
The panel to the left of each record allows navigation through the database. Successive pages can be turned to browse the records. The database can be searched or sorted using any of the data fields. A simple search can be made based on molecular or genetic gene names. Records can be viewed as forms (one record at a time) or as tables (20 records at a time). Further explanation of how to navigate through the database is provided. If links on the record pages do not work it is likely to be because your browser is not enabled to accept "pop-up" windows and you will need to enable "pop-up" windows for this site.
Follow this link for publications from this laboratory describing this work in more detail. This page provides more background on the strategies that have been followed by this laboratory in generating the data in this database.
A screen shot of an example record from the database.